
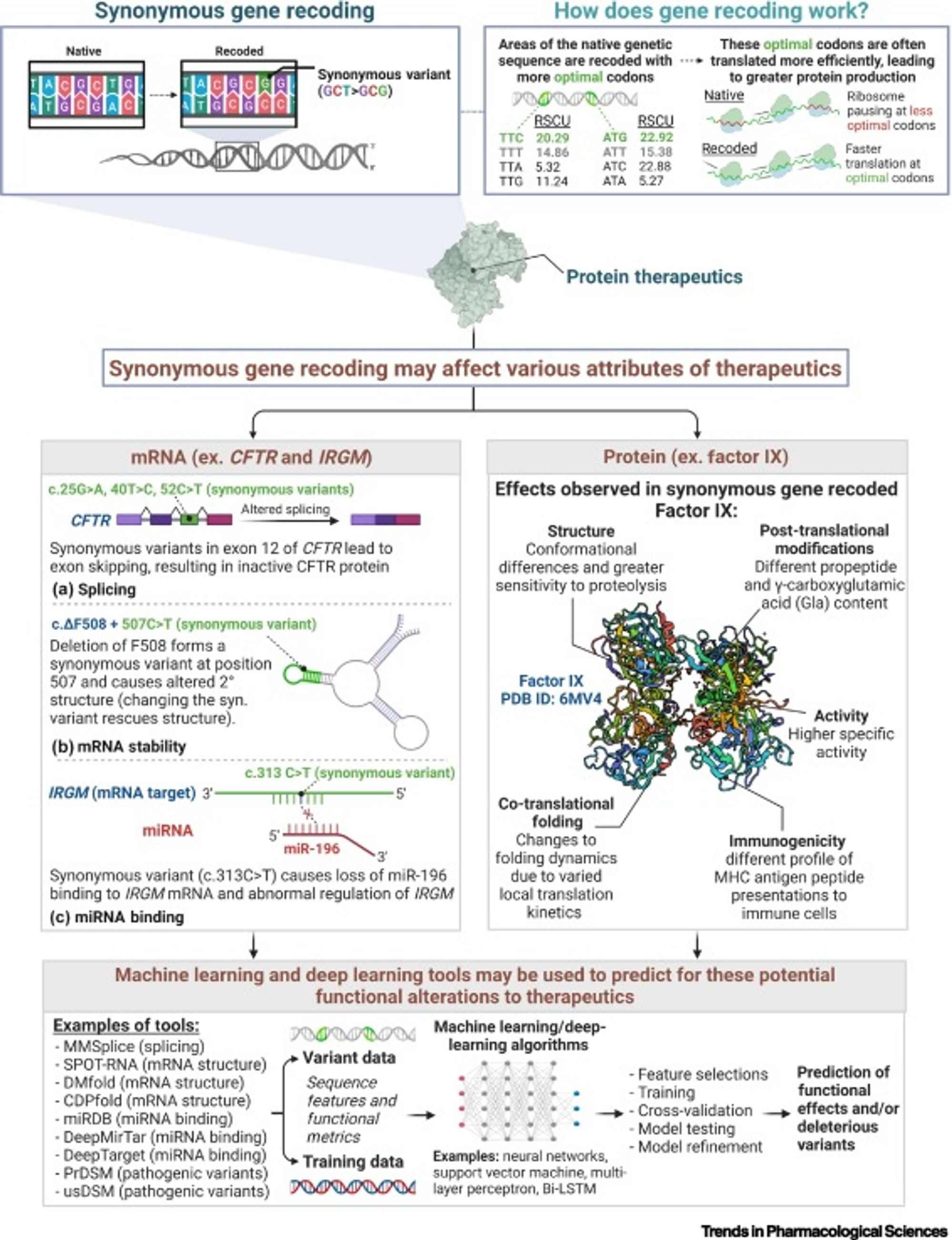
Synonymous gene recoding is a common technique used in gene therapeutic biomanufacturing to improve the production process or efficacy of a targeted therapeutic. The most common example of this approach is synonymous codon substitution, where a three base-pair codon sequence is swapped for another one encoding the same amino acid, resulting in an identical protein sequence. For many gene therapeutics, codon substitution is used to optimize the translation rate of the sequence thus increasing the number of successfully translated protein molecules. Although synonymous codon substitutions do not change the amino acid sequence of the gene therapeutic it can still lead to undesirable and dangerous side-effects. In this blog we discuss a recent publication by Lin et al, on the implications of the negative impacts and the role of artificial intelligence (AI) to overcome these challenges1.
Negative Effects of Synonymous Codon Substitution
Although synonymous codon substitutions do not change the amino acid sequence of the gene therapeutic it can still lead to undesirable and dangerous side-effects. Some potential issues that may arise from synonymous codon substitutions are the following:
- Changes in how messenger RNA transcripts are spliced, driving changes to the final amino acid sequence and subsequent protein expression.
- Changes in the structure and stability of the messenger RNA molecule, leading to faster degradation of the molecule or changes to the rate of translation that negatively impact the quantity of produced protein.
- Impacts on microRNA binding capacity driving a change in the gene regulatory network of the cell.
- Alternative folding of the final protein product, as well as changes in post-translational modifications of the protein, leading to deleterious changes in protein function.
These changes can reduce or eliminate the expression of the therapeutic protein, or alter its functionality and immunogenicity risk through changes to the protein structure.
Leveraging Computational Methods to Optimize Synonymous Codon Substitution and Therapy Production
In a recent opinion article by Lin et al., the authors advocate for use of emerging deep learning methods to help mitigate the negative side effects of synonymous gene recoding in therapeutic development1. There are a number of published neural network models (some of which are enumerated below) to predict the impact of sequence changes on messenger RNA splicing and structure, as well as micro-RNA binding affinity.
MMSplice2 (splicing)
SPOT-RNA3 (mRNA structure)
DMfold4 (mRNA structure)
CDPfold5 (mRNA structure)
miRDB6 (miRNA binding)
DeepMirTar7 (miRNA binding)
DeepTarget8 (miRNA binding)
PredDSM9 (pathogenic variants)
usDSM10 (pathogenic variants)
Using these tools will predict possible deleterious effects of synonymous codon substitutions allowing researchers to screen out unsafe or ineffective codon substitutions. Although we are further away from having a reliable method to predict changes to protein structure due to synonymous changes to its encoding DNA sequence, big recent advancements in the field of protein folding (eg AlphaFold) indicate that there could be promising developments in the near future11.
In the gene therapy space, codon optimization has been tested as a strategy for improving the efficacy of lentiviral vector (LV) gene therapies, yet its application to RD114-TR glycoproteins revealed unexpected roadblocks. Despite increasing expression levels through codon optimization, subsequent modifications such as glycosylation and surface transport were reduced leading to inactive LVs with dramatically reduced potency12. Moreover, codon modification in AAV gene therapy vectors has shown to result in increased CpG content13. Unmethylated CpGs are known to elicit an immune response, which would be desirable in the context of vaccine design, therefore, for the purpose of codon optimization for gene therapy, the reduction of CpG sites is necessary to reduce the immunogenicity of AAV gene therapy13.
To build an optimal strategy in predicting functional synonymous variants, unique features that can identify one algorithm's superiority over another will be key to this process in addition to wet-lab experimental resources. By combining both sets of approaches together, we can create a more comprehensive framework for gaining reliable results in gene therapy development now and into the future.
AI Disclosure: Feature image was generated by an AI image tool.
Stay up-to-date on computational life science research
Sign up for our newsletterReferences
- Brian C. Lin, Nayiri M. Kaissarian, Chava Kimchi-Sarfaty, Implementing computational methods in tandem with synonymous gene recoding for therapeutic development, Trends in Pharmacological Sciences, Volume 44, Issue 2, Pages 73-84 (2023)..
- Cheng, J., Nguyen, T., Cygan, K. et al. MMSplice: modular modeling improves the predictions of genetic variant effects on splicing. Genome Biol 20, 48 (2019).
- GitHub: https://github.com/jaswindersingh2/SPOT-RNA
- Zhang H et al., DMfold: A Novel Method to Predict RNA Secondary Structure with Pseudoknots Based on Deep Learning and Improved Base Pair Maximization Principle. Frontier Genetics Vol 10 (2019).
- GitHub: https://github.com/zhangch994/CDPfold
- Chen Y and Wang X, miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Research, Volume 48, Issue D1, 08 January 2020, Pages D127–D131.
- Wen M.et al.DeepMirTar: a deep-learning approach for predicting human miRNA targets. Bioinformatics. 2018; 34: 3781-3787.
- Lee B.et al. deepTarget: end-to-end learning framework for microRNA target prediction using deep recurrent neural networks. arXiv. 434-442 (2016).
- Li, X et al. PredDSMC: A predictor for driver synonymous mutations in human cancers. Front Genetics Vol 14 (2023).
- Tiejun Zhang T et al, usDSM: a novel method for deleterious synonymous mutation prediction using undersampling scheme. Briefings in Bioinformatics, Volume 22, Issue 5, September (2021).
- Jumper, J., Evans, R., Pritzel, A. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
- Bovolenta C et al, Codon Optimization Leads to Functional Impairment of RD114-TR Envelope Glycoprotein. Molecular Therapy - Methods & Clinical Development, Volume 4, Pages 102-114 (2017).
- J. Fraser Wright, Codon Modification and PAMPs in Clinical AAV Vectors: The Tortoise or the Hare?, Molecular Therapy, Volume 28, Issue 3, (2020).









